Home
Target list
Statistics
Structure
galleries
Community nominated
targets
Technology
development
Web tools and servers
Databases
Participating investigators
Publications
Education and
research training
NESG in the news

salicylic acid-binding protein 2
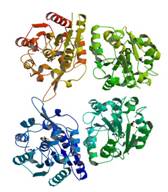
|
X-Ray | Dan Klessig, Cornell University; Boyce Thompson Institute for Plant Research |
PSI-SKB highlight | structure: 1Y7H |
| Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, Klessig DF, Tong L (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci USA, 102: 1773-8. | |
| Zhao N, Guan J, Forouhar F, Tschaplinski TJ, Cheng ZM, Tong L, Chen F (2009) Two poplar methyl salicylate esterases display comparable biochemical properties but divergent expression patterns. Phytochemistry, 70: 37-44. |
antibacterial peptide microcin J25
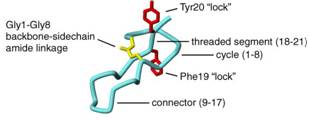 |
NMR | Richard Ebright, Rutgers University | structure: 1PP5 |
| Bayro MJ, Mukhopadhyay J, Swapna GV, Huang JY, Ma LC, Sineva E, Dawson PE, Montelione GT, Ebright RH (2003) Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot. J Amer Chem Soc, 125: 12382-3. |
ribosomal RNA large subunit methyltransferase A
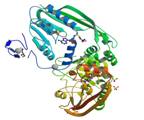 |
X-Ray | Masayori Inouye, UMDNJ | structure: 1P91 |
| Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT (2004) Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc Natl Acad Sci USA. 101: 4041-6 |
lipoprotein ygdR precursor
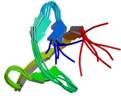 |
NMR | lipoprotein | structure: 2JN0 |
lipoprotein ygdR precursor
 |
X-Ray | lipoprotein | structure: 3FIF |
putative HTH-type transcriptional regulator yddM
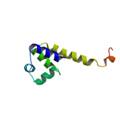 |
X-Ray | PSAM | structures: 2ICP 2ICT |
lipoprotein yehR precursor
 |
NMR | lipoprotein | structure: 2JOE |
uncharacterized lipoprotein yajI
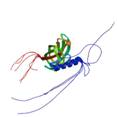 |
NMR | lipoprotein | structure: 2JWY |
lipoprotein spr precursor
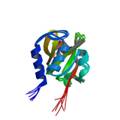 |
NMR | lipoprotein | structure: 2K1G |
| Aramini J, Rossi P, Huang YJ, Zhao L, Jiang M, Maglaqui M, Xiao R, Locke J, Nair R, Rost B, Acton TB, Inouye M, Montelione GT (2008) Solution NMR structure of the NIpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47: 9715-7 |
putative lipoprotein yceB
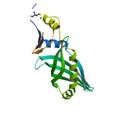 |
X-Ray | lipoprotein | structure: 3EYR |
lipoprotein ymcC precursor
 |
X-Ray | lipoprotein | structure: 2IN5 |
putative outer membrane lipoprtein YiaD
 |
NMR | lipoprotein | structure: 2K1S |
thiamine biosynthesis lipoprotein apbE
 |
X-Ray | lipoprotein | structure: 2O18 |
serine phosphatase of RNA polymerase II CTD
 |
X-Ray | PSAM | structures: 3FDF 3FMV |
human BFL-1 in complex with BAK BH3 peptide
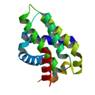 |
X-Ray | Céline Gélinas, UMDNJ | structure: 3I1H |
CAPER RRM2 domain
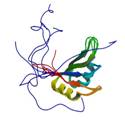 |
NMR | Céline Gélinas, UMDNJ | structure: 2JRS |
kazal-1 Domain of Human Follistatin-related protein 3
 |
NMR | PSAM | structure: 2KCX |
homospermidine synthase
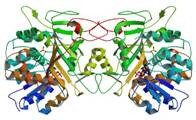 |
X-Ray | Carl Porter, Rosewell Park Cancer Institute, NY | structure: 2PH5 |
LPPG:FO 2-phospho-L-lactate
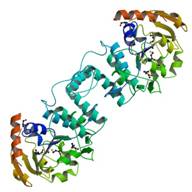 |
X-Ray | Robert White and Ann Galinier, Virginia Tech | structures: 2FFE 3C3D 3C3E 3CGW |
| Forouhar F, Abashidze M, Xu H, Grochowski LL, Seetharaman J, Hussain M, Kuzin A, Chen Y, Zhou W, Xiao R, Acton TB, Montelione GT, Galinier A, White R, Tong L. (2008) Molecular insights into the biosynthesis of the F420 coenzyme. J Biol Chem, 283: 11832-40. |
protein BQLF2
 |
X-Ray | Hongyu Deng and Ren Sun, Chinese Academy of Sciences and UCLA | structures: 2H3R 2OA5 |
CGI121
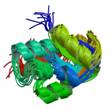 |
NMR | Frank Sicheri, Samuel Lunenfeld Research Institute | structure: 2K8Y |
| Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JS, Szilard RK, Zhang W, Ho CS, Wan L, Fares C, Rumpel S, Kurinov I, Arrowsmith CH, Durocher D, Sicheri F (2008) Atomic structure of the KEOPS complex: an ancient protein kinase-contained molecular machine Mol Cell, 32: 259-75. |
core protein P7
 |
X-Ray | Stuart Firestein, Columbia University | structure: 2Q82 |
DsrR IscA-like protein
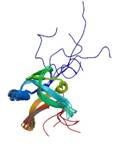 |
NMR | Christiane Dahl and Frauke Grimm, Rheiniche Friedrich-Wilhelms University Bonn, Germany | structure: 2K4Z |
de novo designed ferredoxin-like fold protein
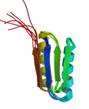 |
NMR | David Baker, University of Washington | structure: 2KL8 |
de novo designed Rossmann 2x2 fold protein
 |
NMR | David Baker, University of Washington | structure: 2KPO |
engineered Lipocalin FluA
 |
NMR | Robert Krug, University of Texas | structure: 1T0V |
N-terminus rat short alpha tropomysin
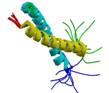 |
NMR | Sarah Hitchcock-DeGregori, UMDNJ | structure: 1IHQ |
nonstructural protein NS1
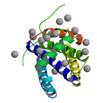 |
X-Ray | Robert Krug, University of Texas | structure: 1XEQ |
NS1A
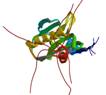 |
NMR | Robert Krug, University of Texas | structure: 2KKZ |
NS1A
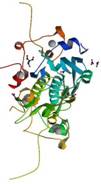 |
X-Ray | Robert Krug, University of Texas | structure: 2RHK |
| Yin C, Khan JA, Swapna GV, Ertekin A, Krug RM, Tong L, Montelione GT (2007) Conserved surface features form the double stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses.J Biol Chem 282: 20584-92. |
tropomyosin 1 alpha chain
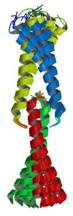 |
NMR | Sarah Hitchcock-DeGregori, UMDNJ | structures: 2G9J 1MV4 |
| Greenfield N, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE (2006) Solution NMR structure of the junction between tropomysin molecules: implications for actin binding and regulation J Mol Biol 364: 80-96. | |
| Greenfield N, Swapna GV, Huang YJ, Palm T, Graboski S, Montelione GT, Hitchcock-DeGregori SE (2003) The structure of the carboxyl terminus of striated alpha-tropomysin in solution reveals an unusual parallel arrangement of interacting alpha-helices Biochemistry 42: 614-9. |
protein yktB
 |
X-Ray | MCSG | structure: 2A8E |
protein yndB START domain
 |
NMR | MCSG | structure: 2KEW |
 |
NMR | MCSG | structure: 2HFI |
protein yppE
 |
X-Ray | MCSG | structure: 2IM8 |
 |
NMR | MCSG | structures: 1XN8 1ZTS |
protein ysnE
 |
NMR | MCSG | structure: 1YX0 |
protein yutD
| NMR | MCSG | structure: 1KL5 |
protein YvbK
 |
X-Ray | MCSG | structure: 1YVK |
yxeF precursor protein
 |
NMR | lipoprotein | structure: 2JOZ |
uncharacterized protein yobA
 |
NMR | lipoprotein | structure: 2JQO |
uncharacterized lipoprotein yjhA
 |
X-Ray | lipoprotein | structure: 3CFU |
probable amino-acid ABC transporter
 |
X-Ray | lipoprotein | structure: 2O1M |
probable ABC transporter
 |
X-Ray | lipoprotein | structure: 2IEE |
iron(III)-hydroxamate-binding
 |
X-Ray | lipoprotein | structure: 3HXP |
ferrichrome-binding protein
 |
X-Ray | lipoprotein | structure: 3G9Q |
iron-uptake system-binding protein
 |
X-Ray | lipoprotein | structure: 2PHZ |
protein YcdH
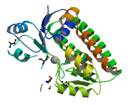 |
X-Ray | lipoprotein | structure: 2O1E |
YerB protein
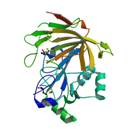 |
X-Ray | lipoprotein | structure: 2PSB |
protein yxiM precursor
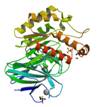 |
X-Ray | lipoprotein | structure: 2O14 |
protease synthase and sporulation
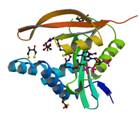
|
X-Ray | Carl Porter, Roswell Park Cancer Institute, NY | structure: 1TIQ |
| Forouhar F, Lee IS, Vujcic J, Vujcic S, Shen J, Vorobiev SM, Xiao R, Acton TB, Montelione GT, Porter CW, Tong L (2005) Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase J Biol Chem, 280: 40328-36. |
virulence protein STM3117
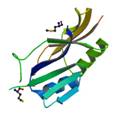
|
X-Ray | Liang Shi, PNNL, WA | structure: 3HNQ |
VCA0042 complexed with c-di-GMP
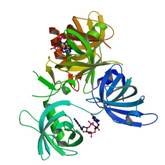
|
X-Ray | Andrew Camilli, Tufts MA | structure: 2RDE |
| Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF (2007)The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO, 26: 5153-66. |
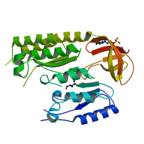
|
X-Ray | Marc Fontecave and Hamid Atta, CEA, Grenoble France | structure: 2QGQ |
Tryptophan deoxygenase
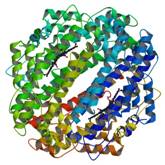
|
X-Ray | Stephen Chapman, University of Edinburgh, Scotland | structure: 2NW7 |
| Forouhar F, Abashidze M, Xu H, Grochowski LL, Seetharaman J, Hussain M, Kuzin A, Chen Y, Zhou W, Xiao R, Acton T, Montelione GT, Galinier A, White RH, Tong L (2008) J Biol Chem 283: 11832-40. |
UDP-N-acetylglucosamine tranferase

|
NMR | Barbara Imperiiali, MIT, MA | structure: 2JZC |
| Wang X, Weldeghiorghis T, Zhang G, Imperiali B, Prestegard JH (2008) Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase. Structure 16:965-75. |
